Timeseries
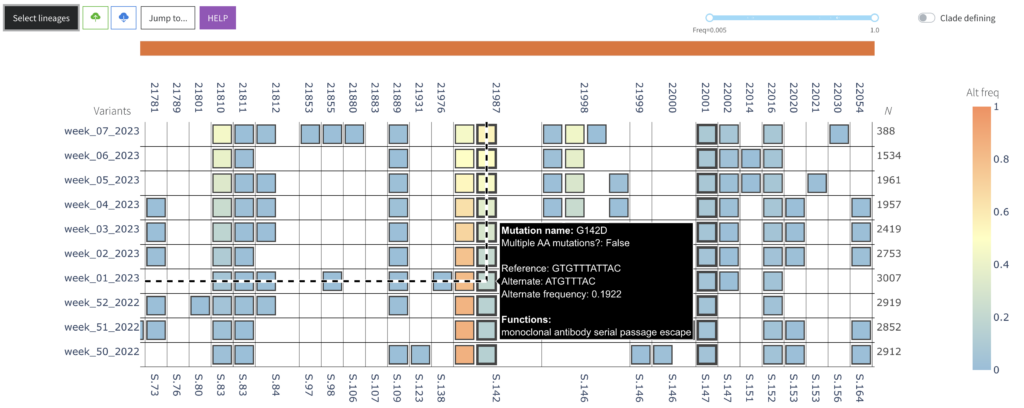
VIRUS-MVP vizualization is designed to be data-agnostic, allowing visualisation of viral sequences that can be grouped by any specified variable.
For example, sequences can be sorted by a period of time (default: 7 days) in order to identify the relative increase or decrease in mutation prevalence.
Below is a link to an interactive demo of the SARS-CoV-2 time series data.
Annotated SLURM Script to generate time series data
#!/bin/bash #SBATCH --time=2:00:00 #SBATCH --job-name=nf-ncov-voc-timeseries #SBATCH --cpus-per-task=4 #SBATCH --nodes=2 #SBATCH --mem-per-cpu=12G #SBATCH --error=%x-%j.error #SBATCH --output=%x-%j.out module load nextflow module load apptainer nextflow run main.nf \ -profile singularity \ --prefix <prefix> \ --grouping_criteria time \ --mode reference \ --skip_pangolin \ --skip_nextclade \ --startdate <yyyy-mm-dd> \ --enddate <yyyy-mm-dd> \ --outdir <results_dir>
